PCR laboratory testing, which became a household phrase during the Covid pandemic, is being used in the first stages of a regulatory case study project to register a viral bioinsecticide.
The A Lighter Touch project led by programme partners Vegetables NZ and Key Industries Ltd is aiming to register the product for use against diamondback moth (DBM).
One of the first steps is to establish whether or not the virus (PlxyGV), the active ingredient in the viral bioinsecticide, is already present in New Zealand. This will be done by using a quantitative polymerase chain reaction (qPCR) assay, or test, to show if the virus is present in New Zealand DBM caterpillars.
Over the last three summers, about 500 diamondback moth larvae have been collected in multiple regions and stored in freezers. DNA extracted from these frozen caterpillars will now be analysed to see if it includes the DNA of the PlxyGV virus.
The process is illustrated through photographs showing the ‘inputs’ and ‘outputs’ of the qPCR assay. Figure 1 (below) shows the ‘input’, small test tubes in which DNA is being extracted from individual caterpillars at the dnature laboratory in Gisborne.

Figure 1: qPCR ‘input’ – DNA extraction of frozen DBM caterpillars.
A small volume of the liquid in the tube is then analysed for the presence of PlxyGV DNA. It is detected by increasing fluorescence as the DNA is amplified up in repeated cycles of the PCR reaction.
dnature have developed a qPCR assay which simultaneously looks for two pieces of the virus DNA. An internal control is also run – this looks for a piece of non-virus DNA which is already in the sample and confirms that the DNA amplification process is working correctly.
In addition, negative and positive controls for the virus DNA are run. The negative control has no DNA added so for a valid result it should show no amplification (i.e. no contamination present).
As there is no PlxyGV virus known in NZ, a synthetic positive control has had to be made by dnature. The DNA of PlxyGV has been sequenced overseas and from that information, part of the viral DNA has been synthesised in the laboratory for use as the positive control.
An example of the ‘output’ of the qPCR assay is shown in Figure 2 (below) where there are three graphs plotting fluorescence versus PCR reaction cycles. In the top two graphs, the results for each of the pieces of the virus DNA are shown. The graph curves show the dilutions of the positive control: the lower the number of cycles before the curve phase starts, the greater the virus concentration present.
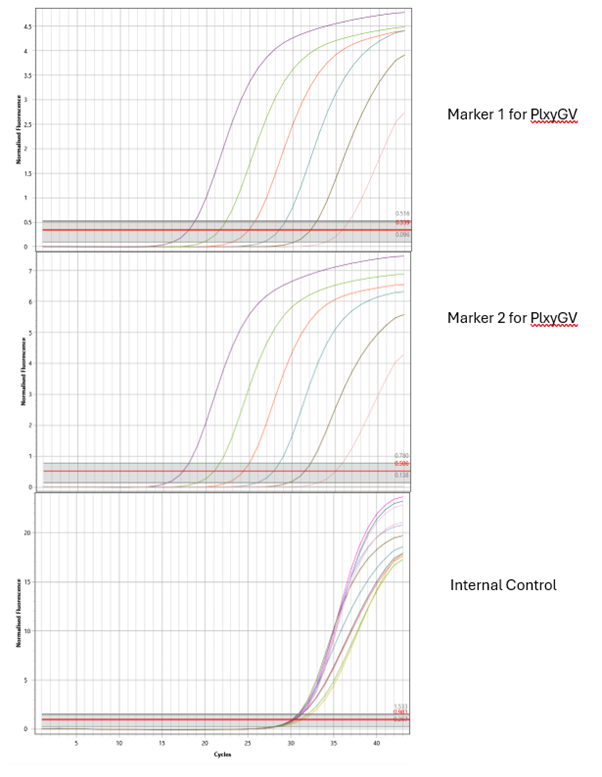
Figure 2: qPCR ‘output’ – results from testing DNA extracted from one DBM caterpillar.
One of the horizontal lines at the bottom of each graph represents the negative control (showing no DNA amplified – as expected for a good assay run). The second horizontal line is for the DNA extracted from a caterpillar which means that in this test neither of the target sequences of PlxyGV were found in the sample.
Lastly, the bottom graph in Figure 2 shows the amplification of the internal control DNA showing that good quality DNA was added to the reactions.
So far, samples from a few dozen of the frozen caterpillars have been run to test the process. None of the samples tested so far have shown the PlxyGV virus to be present. The results for the remaining tests will determine next steps for the project.
If the virus is found in the remaining samples, that will be part of the evidence needed to support a case for an application for the PlxyGV virus to be declared ‘not new’ to New Zealand.
However if sample testing does not find any evidence of the virus, then the next step will be to make an application for the virus to be imported as a New Organism under containment for host range testing against DBM and other possible hosts in New Zealand.
